| Bos taurus Gene: UBE2I | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||
| InnateDB Gene | IDBG-634261.3 | ||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||
| Gene Symbol | UBE2I | ||||||||||||||||||||
| Gene Name | SUMO-conjugating enzyme UBC9 | ||||||||||||||||||||
| Synonyms | |||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000038866 | ||||||||||||||||||||
| Encoded Proteins |
SUMO-conjugating enzyme UBC9
|
||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||
| InnateDB Annotation from Orthologs | |||||||||||||||||||||
| Summary |
[Mus musculus] Baiap2l1 recruits Ube2i to sumoylate Pcbp2, which causes its cytoplasmic translocation during viral infection and the sumoylated Pcbp2 associates with Mavs to initiate its degradation, leading to downregulation of antiviral responses.
|
||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000103275:
The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation. Ubiquitination involves at least three classes of enzymes: ubiquitin-activating enzymes, or E1s, ubiquitin-conjugating enzymes, or E2s, and ubiquitin-protein ligases, or E3s. This gene encodes a member of the E2 ubiquitin-conjugating enzyme family. Four alternatively spliced transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Jul 2008] |
||||||||||||||||||||
| Gene Information | |||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||
| Genomic Location | Chromosome 25:1041085-1048106 | ||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||
| Band | |||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||
| Interactions | |||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 280 interaction(s) predicted by orthology.
|
||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||
| Orthologs | |||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||
| NETPATH |
AndrogenReceptor pathway
TGF_beta_Receptor pathway
TNFalpha pathway
|
||||||||||||||||||||
| REACTOME |
Meiotic synapsis pathway
Post-translational protein modification pathway
Processing and activation of SUMO pathway
Cell Cycle pathway
Metabolism of proteins pathway
SUMO is transferred from E1 to E2 (UBE2I, UBC9) pathway
SUMOylation pathway
Meiosis pathway
Meiotic Synapsis pathway
Mus musculus biological processes pathway
SUMOylation pathway
Meiotic synapsis pathway
Cell Cycle pathway
Metabolism of proteins pathway
Post-translational protein modification pathway
SUMO is transferred from E1 to E2 (UBE2I, UBC9) pathway
Processing and activation of SUMO pathway
Meiosis pathway
|
||||||||||||||||||||
| KEGG |
Ubiquitin mediated proteolysis pathway
RNA transport pathway
Ubiquitin mediated proteolysis pathway
RNA transport pathway
|
||||||||||||||||||||
| INOH | |||||||||||||||||||||
| PID NCI |
Coregulation of Androgen receptor activity
Signaling events mediated by HDAC Class I
Signaling events mediated by HDAC Class II
C-MYB transcription factor network
Sumoylation by RanBP2 regulates transcriptional repression
Regulation of cytoplasmic and nuclear SMAD2/3 signaling
|
||||||||||||||||||||
| Cross-References | |||||||||||||||||||||
| SwissProt | |||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||
| UniGene | Bt.46371 Bt.86966 | ||||||||||||||||||||
| RefSeq | NM_001099372 | ||||||||||||||||||||
| HUGO | |||||||||||||||||||||
| OMIM | |||||||||||||||||||||
| CCDS | |||||||||||||||||||||
| HPRD | |||||||||||||||||||||
| IMGT | |||||||||||||||||||||
| EMBL | |||||||||||||||||||||
| GenPept | |||||||||||||||||||||
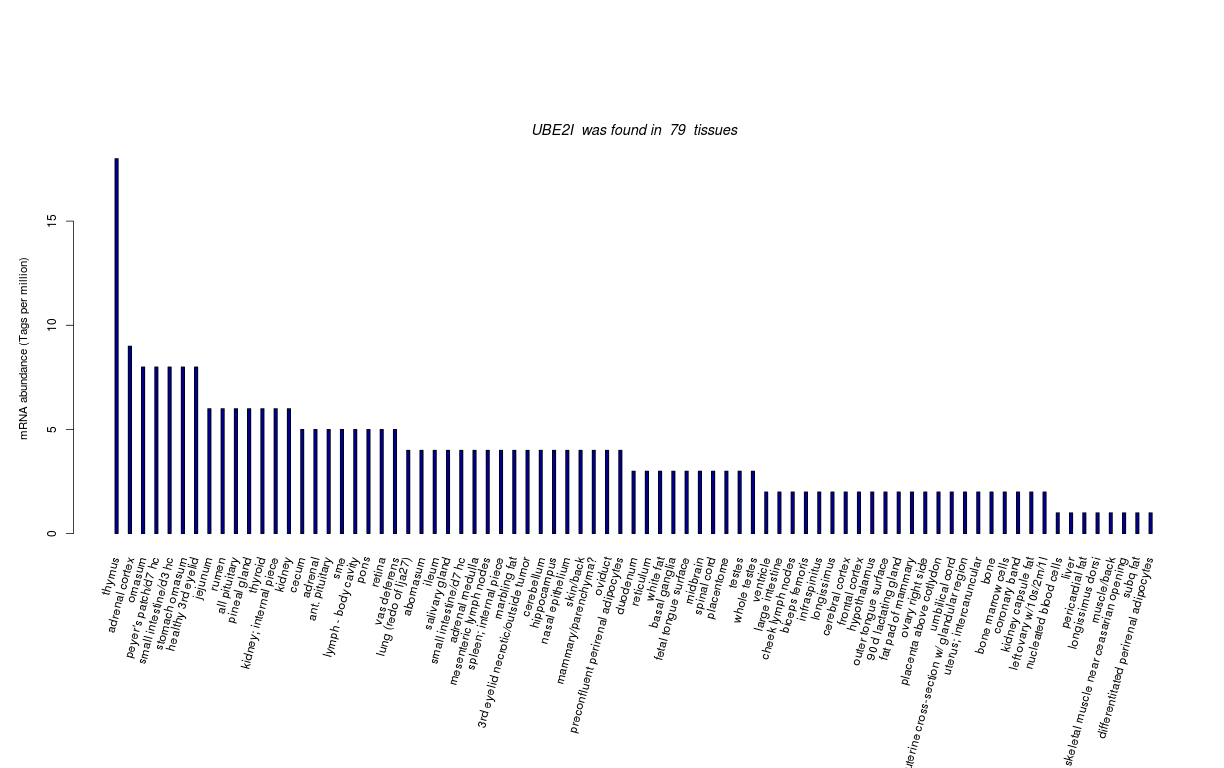
| RNA Seq Atlas | |||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||